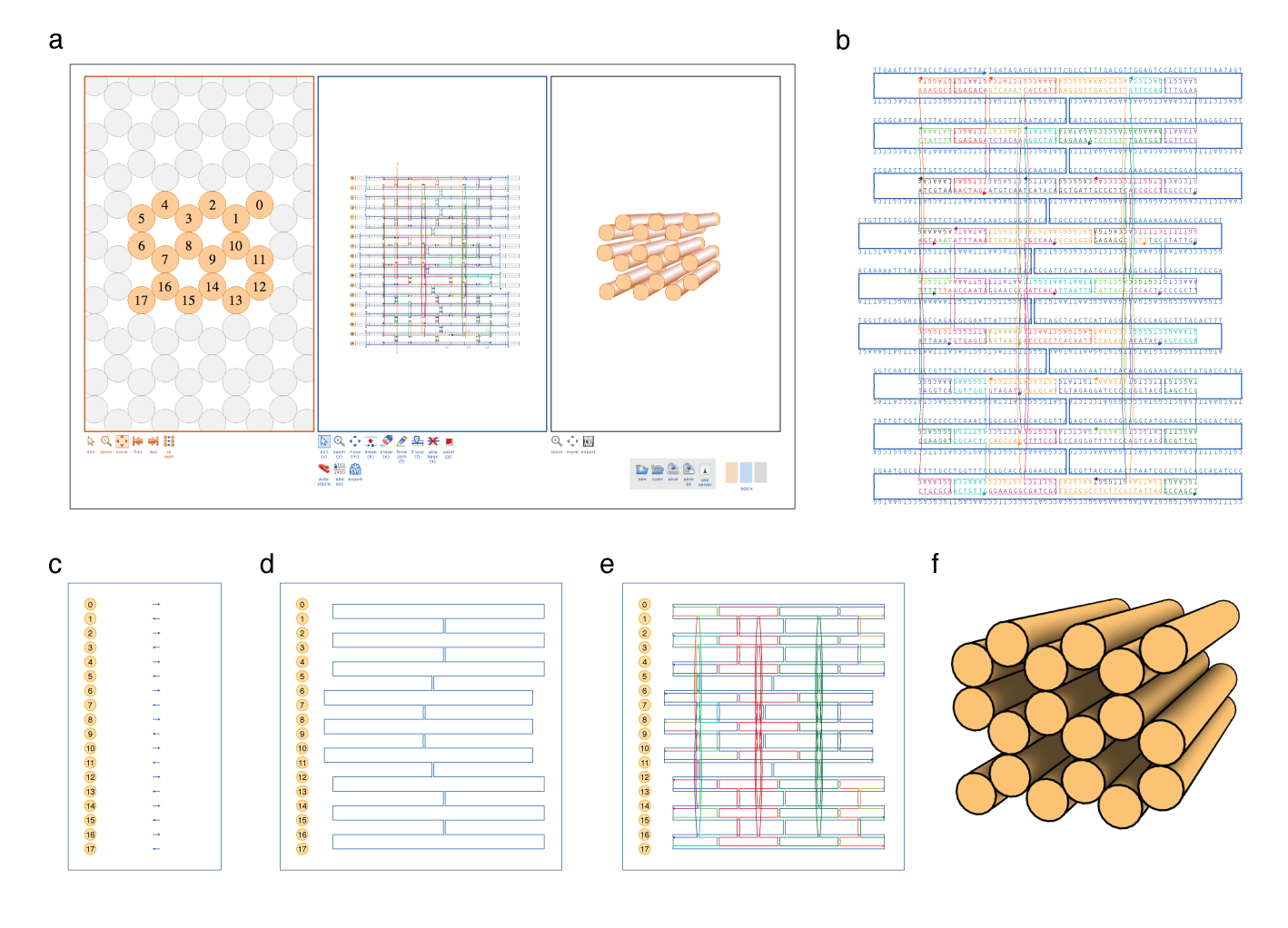
Vertical grid lines are drawn thicker near default staple crossovers every 7 bases for honeycomb lattice and 8 bases for square lattice. Preparing an input design file using Tiamat In this tutorial, we will use the. This part of the assignment is slightly independent from the caDNAno one. New selection-based workflow The interface for editing strands has been significantly changed. The color of each strand in the atomic model is the same as in the caDNAno design. 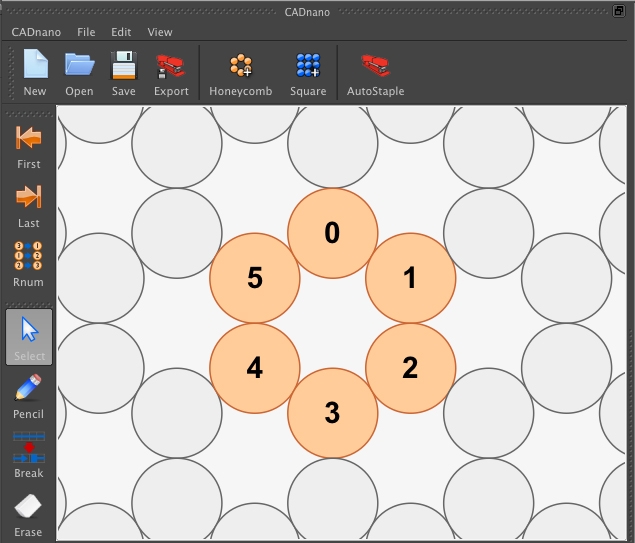
| Uploader: | Mauktilar |
| Date Added: | 12 September 2009 |
| File Size: | 39.82 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 59865 |
| Price: | Free* [*Free Regsitration Required] |
Week 2. DNA Nanostructures
No more erase tool Selected objects are removed by pressing the delete key. Here is the link to this week assignments candano. CanDo analysis must be performed at the fine model resolution to generate the atomic model.
It is not recommended to use a nick stiffness factor less than the default value as it may result in much slower or no convergence of the analysis.
Alternatively, users may enter their own values.
cadnano version 2.1
Selection filters In order to allow precise selections, we also have added selection filters. This will update caDNAno.
Do not give up. If you leave the Added "about cadnano" dialog. Once the CanDo analysis is completed, users will receive results in ZIP files attached in separate emails sent to the email address they filled in the submission form.
Automatic scaffold creation Cadnano will attempt to "pre-populate" some scaffold strands when you click and drag in the lattice panel. Cadnnano undo to revert to the 3-base stubs.
Tutorial – CanDo
Browse to the location of your Tiamat sequence file. Sequence labels have been moved "inside" the strand grid in the path view. The next step is to convert the Pacman pattern obtained before into a list of pipetting instructions. I hope you have good imagination now.
In the Command Prompt window of a Windows platform, run the following command. Zoom level of detail: They will snap to "legal" positions.
In a terminal window, navigate to the folder where you want the caDNAno folder to be placed and type:. Choose whether movies visualizing thermal fluctuations of the solution shape will be included with the results.
After stabilizing the data model in the 2.
Lattice-based design caDNAno 1. Viewing the results Once the CanDo analysis is completed, users will receive results in ZIP files attached in carnano emails sent to the email address they filled in the submission form.
But I was getting some errors, so I raised an issue to github and the developers updated the master branch, which solved the problem If something does not work find out what's the problem. Browse to the location of your caDNAno design file. User information provided will be used only for internal evaluation of the resource by counting the number of unique users and the number of unique affiliations.
So I decided to go with caDNAno 2. CanDo offers a new option for users to generate an atomic model of the solution shape of DNA nanostructure.
cadnano version | cadnano
The second character is the row number of the 96 well-plate from A to H. For example, the command echo ABCxyz The color of each strand in the atomic model is the same as in the caDNAno design. If the atomic model is to be included in the results, make sure that the fine resolution is selected in step e.
It had slowed down significantly compared to cadnano1 due to the new data model architecture. Users have an option to use the fine model resolution that computes the shape and flexibility at a cqdnano basepair resolution. Inside that folder cd cadnano2.

Комментариев нет:
Отправить комментарий